Introduction
The WingX project is an RTD (Research, Technology and Development) project was started in 2008 and is funded by SystemsX.ch – the Swiss Initiative in Systems Biology. The principal leader of WingX Prof. Ernst Hafen (ETH Zurich) brings biologists, computer specialists, mathematicians and engineers from 15 research groups from the ETH Zurich, Universities of Zurich, Bern and Basel, and EPF Lausanne together to work on the development of the Drosophila wing at the systems biology level.
In that context, the goal of this project is the implementation of a computational framework to reverse engineer gene regulatory networks controlling morphogenesis of multi-cellular systems from gene expression data (mRNA levels).
GeneNetWeaver (GNW)
Over the last decade, numerous methods have been developed for inference of regulatory networks from gene expression data. However, accurate and systematic evaluation of these methods is hampered by the difficulty of constructing adequate benchmarks and the lack of tools for a differentiated analysis of network predictions on such benchmarks.
We developed a novel and comprehensive method for in silico benchmark generation and performance profiling of network inference methods available to the community as an open-source software called GeneNetWeaver (GNW). In addition to the generation of detailed dynamical models of gene regulatory networks to be used as benchmarks, GNW provides a network motif analysis that reveals systematic prediction errors, thereby indicating potential ways of improving inference methods.The accuracy of network inference methods is evaluated using standard metrics such as precision-recall and receiver operating characteristic (ROC) curves. We used GNW to provide the international DREAM (Dialogue for Reverse Engineering Assessments and Methods) competition with three network inference challenges (DREAM3, DREAM4, and DREAM5).
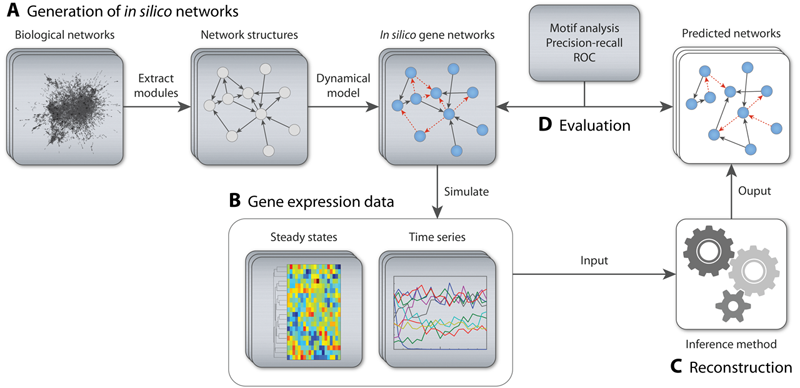
- GeneNetWeaver: In silico benchmark generation and performance profiling of network inference methods (Schaffter et al., 2011). Watch the movie in HD and fullscreen on YoutTube.
Softwares released
 |
GeneNetWeaver (GNW) Automatic generation of realistic benchmarks and performance profiling of genetic network inference methods. Developers: Thomas Schaffter, Daniel Marbach, Gilles Roulet Technology: Java Web Start [website] [manual] [screencast] [launch] Version 3.0.1 |
 |
libSDE libSDE is a Java library for numerical integration of Stochastic Differential Equations (SDEs). Developer: Thomas Schaffter Technology: Java [website] [manuscript] [download] Version 1.0.1 with example |
Main Publications
Journal Articles
Fluorescence Behavioral Imaging (FBI) tracks identity in heterogeneous groups of Drosophila
PLOS One. 2012. Vol. 7, num. 11, p. e48381. DOI : 10.1371/journal.pone.0048381.GeneNetWeaver: In silico benchmark generation and performance profiling of network inference methods
Bioinformatics. 2011. Vol. 27, num. 16, p. 2263-2270. DOI : 10.1093/bioinformatics/btr373.Revealing strengths and weaknesses of methods for gene network inference
PNAS. 2010. Vol. 107, num. 14, p. 6286-6291. DOI : 10.1073/pnas.0913357107.Generating Realistic In Silico Gene Networks for Performance Assessment of Reverse Engineering Methods
Journal of Computational Biology. 2009. Vol. 16, num. 2, p. 229-239. DOI : 10.1089/cmb.2008.09TT.Reports
Numerical Integration of SDEs: A Short Tutorial
2010
Stochastic Simulations for DREAM4
2009